by Caitlin Lynch (LaGCC, Environmental Science, 2022-2023 CRSP cohort)
The work was done as a part of the CRSP program at LaGuardia Community College/CUNY, under the supervision of Dr. Lucia Fuentes.
This article has been published as part of the Special Edition of Ad Astra, which features the CUNY Research Scholars Program (CRSP) across The City University of New York. The issue is accessible at http://adastraletter.com/2024/crsp-special-edition/.
ABOUT THE AUTHOR

Caitlin Lynch
Caitlin Lynch is currently a graduate student at Brooklyn College in the Earth and Environmental Sciences program. Previously, she obtained an Associate’s degree from LaGuardia Community College in the Environmental Science program where she found a passion for fieldwork, research and wetland restoration. During her time at LaGCC she participated in the CRSP research program as well as an internship with the Newtown Creek Alliance supporting their intertidal habitat projects. She is currently working with the Science and Resiliency Institute of Jamaica Bay collecting and analyzing field data. When she is not knee deep in the mud, she enjoys riding her bike around the city, volunteering at beach clean-ups and learning to identify as many plants as possible.
New York City’s East River is a highly anthropogenically affected tidal estuary that is home to many diverse species of plants, animals and microorganisms. Shoreline development of recreational and greenspaces has increased in the past decade, providing a unique opportunity to study the environmental impacts of different types of restoration projects. Considering that marine sediments contain high levels of bacterial diversity and play an essential role in nutrient cycling, degradation of pollutants, as well as being contaminants themselves, profiling these microbiomes can provide insights into the overall ecological/ecosystem health and guidance for restoration planning (Córdova-Kreylos 2006, NYS DEC 2014). The objective of this study is to compare bacterial classes and diversity between three shoreline sites and evaluate whether differences in bacterial diversity indicate varied levels of ecosystem health. We hypothesized that the bacterial communities will differ based-on the type of restoration, with the highest diversity being at the site designed to function like a tidal marsh. Sediment samples and water samples were collected from three sites: Hunters Point Park, Bushwick Inlet Park and Newtown Creek. DNA purification of the samples was processed at the LaGuardia Community College (CUNY) biology lab using the MoBio DNA isolation techniques and sent to an off-campus lab for metagenomic analysis using 16S rRNA sequencing. Ancillary environmental conditions were measured, including water pH, water temperature, dissolved oxygen and carbon dioxide, nitrates, phosphates, and salinity. Our results showed that the environmental conditions among sites showed had little variation in pH, salinity, and temperatures, but a wide range of dissolved oxygen levels. We found that the sediment bacteria reflected unique biochemical processes at each site. The alpha diversity was greater than the beta diversity, and each site had a unique profile of classes with Gammaproteobacteria being the most represented across all 3 sites. Hunter’s Point Park was the site with the greatest bacterial diversity by class. We think that this type of study can be useful as a longitudinal research project to help inform policy makers and stakeholders, especially as local community and advocacy groups become interested in protecting coastal habitats.
New York City’s East River is a tidal estuary highly affected by anthropogenic activity and home to many diverse species of plants, animals and microorganisms (Wigand 2014). Development of recreational and greenspaces along the east side of the river (Brooklyn and Queens) has ranged from bulkhead walls to tidal marshes and riprap and sandy beaches, which provides an opportunity to study the environmental impacts and contributions of these different restoration projects. While many organisms can be used as biomarkers for ecosystem health, bacteria and, specifically, sediment bacteria constitute some of the most important microbes essential for nutrient cycling, degradation of pollutants, and contaminant indicators (Madsen 2011, Wood 2021). Therefore, profiling the bacterial diversity of sediments using metagenomics can provide insights into the overall ecological health and ecosystem services of the restoration project as well as potential guidance for future land-use and conservation planning.
Bacteria are essential components to all ecosystems and organisms on Earth for their metabolic capacities that allow for the breakdown, storage and release of nutrients that would otherwise be trapped in the environment, such as nitrogen, phosphorus and carbon (Madsen 2011, DWER 2023). In tidal estuaries like the New York Harbor and other wetlands such nutrient cycling occurs between the water and sediments. Common urban pollutants such as hydrocarbons, heavy metals and QACs can enter the water and settle on the sediments making the sediment layer highly indicative of the inorganic composition of the marine environment (Mesa 2016). Healthy ecologically functioning coastal systems are able to remove excess organic nitrogen from the water, store organic carbon, and in some instances remove toxins, which are all processes mediated by sediment microbes and the flux of tides that creates oscillating anoxic and oxic conditions (Barbier 2011). In addition, coastal wetlands are sites of high rates of primary productivity, water purification and coastal protection (Kennish 2001).
These important coastal areas are at risk of destruction due to the effects of climate change (sea level rise and storm surge) and increased development (habitat loss, biodiversity loss, pollution) (Kennish 2001). Many managers and planners are implementing green infrastructure projects as a strategy to combat these impacts (Palinkas 2022). One such area is New York, with both increased development and wetland restoration along its shorelines. We chose to evaluate three closely located sites (Figure 1) based on their different restoration types and land use status. Bushwick Inlet Park (Site 1) is a recently restored park with a rocky shoreline primarily designed for human recreational use. Hunter’s Point Park (Site 2) is a shoreline park designed to function like a tidal marsh with areas planted with native salt-tolerant flora that are inundated with the tide. Newtown Creek (Site 3) is a mostly unrestored Superfund site with bulkhead walls and concrete steps with water access. These three areas present an opportunity to measurably compare the microbial biodiversity present in these different restoration designs. Our study asks the following question: Do the alpha and beta diversity of sediment bacteria vary between different shoreline sites restored using different strategies and, if so, do these differences indicate varied levels of ecosystem health? We hypothesized that due to the increased natural areas, tidal flooding, plant and root interactions, Site 2 would have the highest microbial and metabolic diversity. In addition, due to high levels of legacy pollution and little/no vegetation or growth mediums, Site 3 would have the lowest diversity.
Two sediment and water samples were collected from Sites 1 and 3 and three samples from Site 2 on the same day, time, weather conditions and at low tide for access to sediment. All samples were collected in the winter of 2022. Sediment samples were taken at the surface (0-5 cm), collected in sterile plastic containers, transported below 4°C and then stored in a freezer in the lab. Water samples were taken in the surface water above the collected sediment area in sterile bottles. The MoBio soil and water DNA isolation procedures were used to extract the microbial DNA from sediment and water samples, which were stored at -20°C. DNA extracted from the samples were sent to an off-campus lab for metagenomic analysis. Environmental conditions around sampling sites were also measured, including water pH, water temperature, dissolved oxygen (at time of collection), nitrates, phosphate, salinity, and quaternary ammonium compounds (QAC). Dissolved oxygen was measured using the LaMotte Dissolved Oxygen Water Test Kit and the other measurements were taken using their corresponding Hach Test Kits.
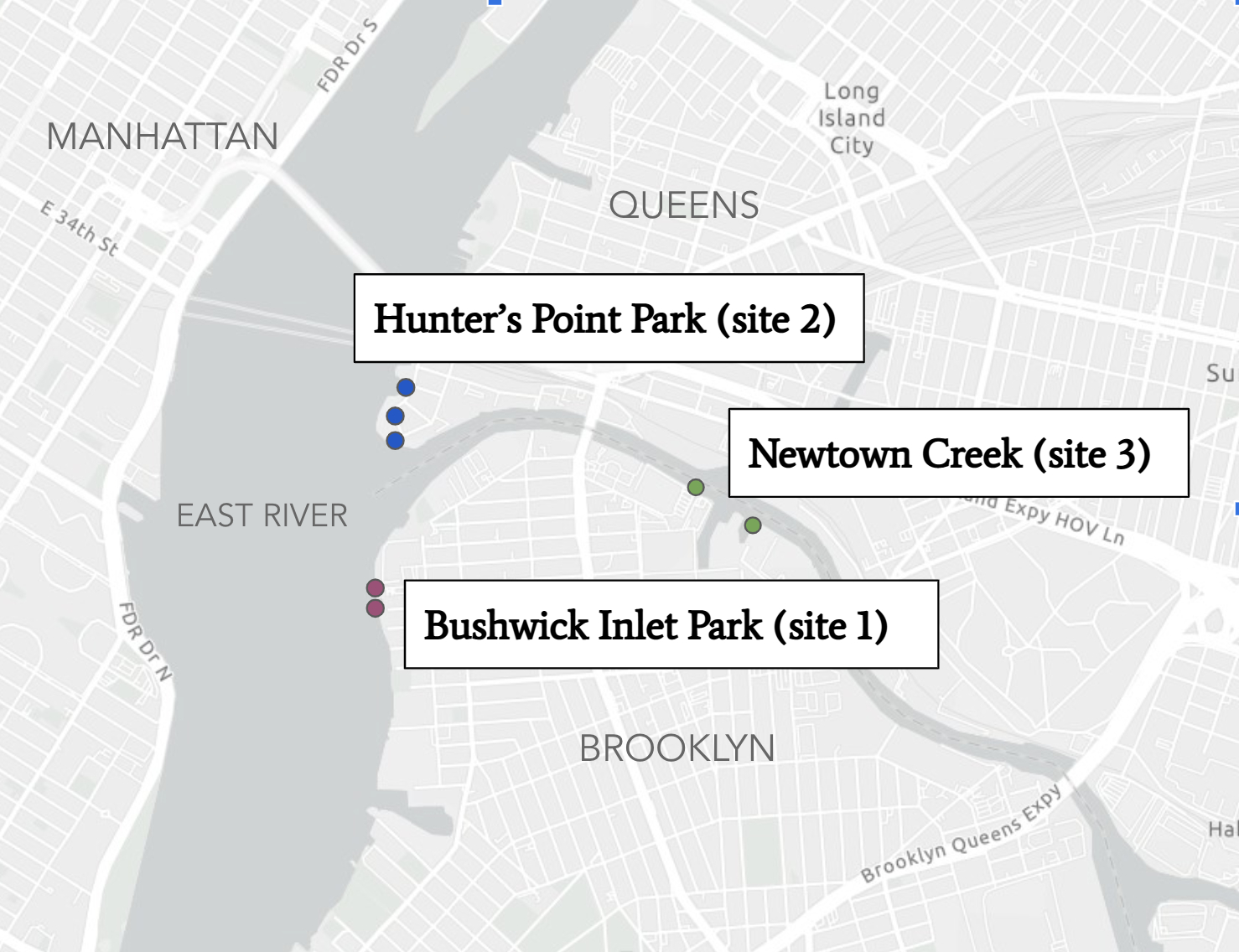
Figure 1: Site locations of data collection.
The data collected reflects that each site contains a different bacterial profile, in both diversity and abundance. We found that the alpha diversity was greater than the beta diversity, and each site had a unique profile of bacterial classes. While just a few classes (Gammaproteobacteria, Deltaproteobacteria, and Flavobacteriia) represent the largest groups at each location, the relative abundance of these differs as well as the diversity of other classes present. Hunters Point Park had the highest class diversity with 16 classes detected, followed by Newtown Creek with 11 classes and Bushwick Inlet Park with 9 classes. Between 63.77%–90.45% of the microbes were defined as Unclassified.
The largest portion by class in all samples was Gammaproteobacteria, which is characteristic of marine sediments, and reflective of this large diverse class that also includes many known pathogens, such as E. coli and Salmonella (Wood 2021). Sites 2 and 3 shared the second largest class, Deltaproteobacteria, yet Site 1 showed a much smaller portion of this class. Unique to Site 1 was a large portion of the phyla Saprospirae, which includes the genus Cytophaga, important cellulose decomposers and remineralizers of organic materials, and Beggiatoa, key in sulfur respiration. Unique to Site 2 was Anaerolineae, a common marine sediment bacterium in anaerobic environments, suggesting lower oxygen levels in the sediment. Site 1 contained a unique prevalence of Loktanella, Winogradskyella and Glaciecola, all newly identified marine organisms. Site 3 contained a high prevalence of sulfur-reducing bacteria with high counts of Sulfurimonas, Desulfococcus, Desulfosarcina, not present in other sites indicating sulfate reduction without oxygen. Site 2 contained a small amount of a huge range of different genera with no defined groups over 1% of the total sample.
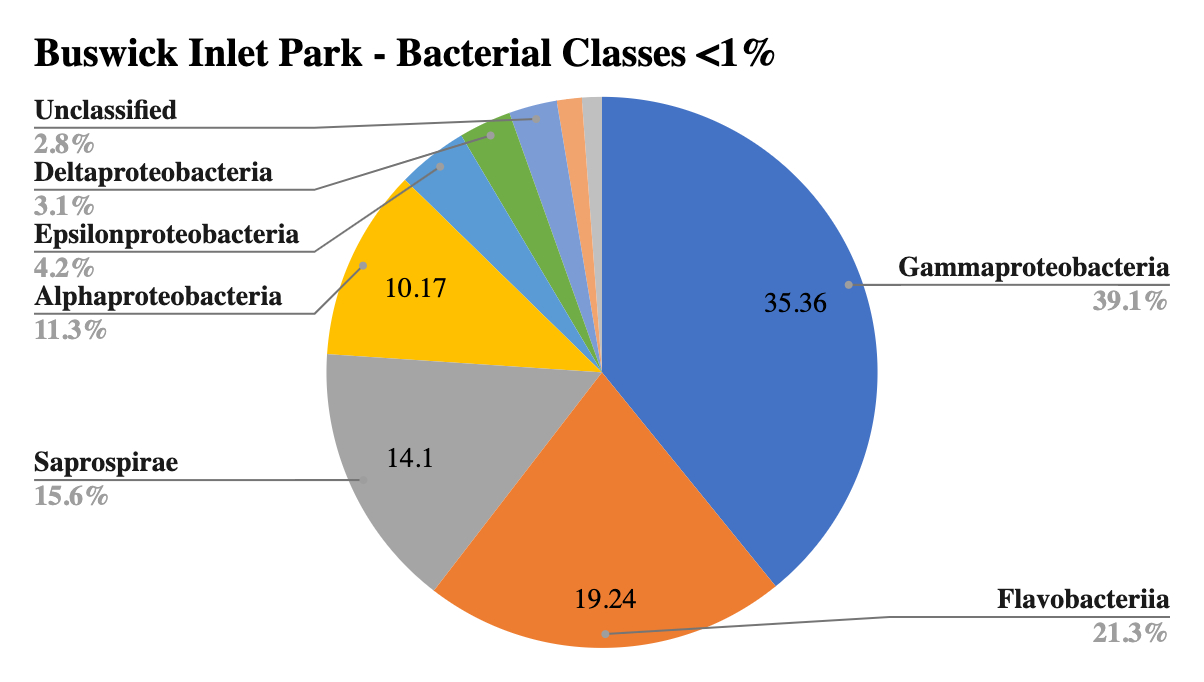
Figure 2: Bacterial diversity by class at Bushwick Inlet Park.
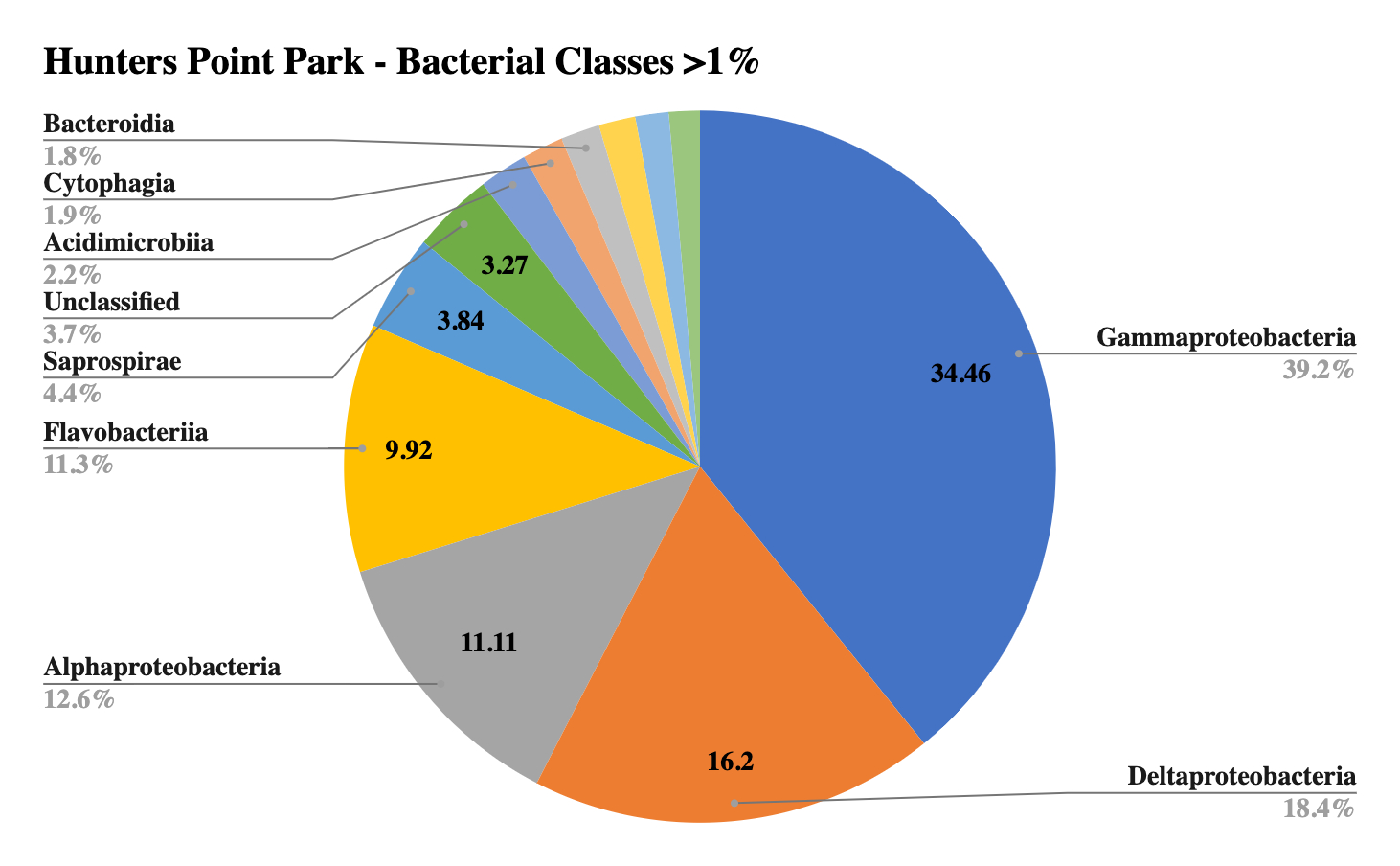
Figure 3: Bacterial diversity by class at Hunter’s Point Park.
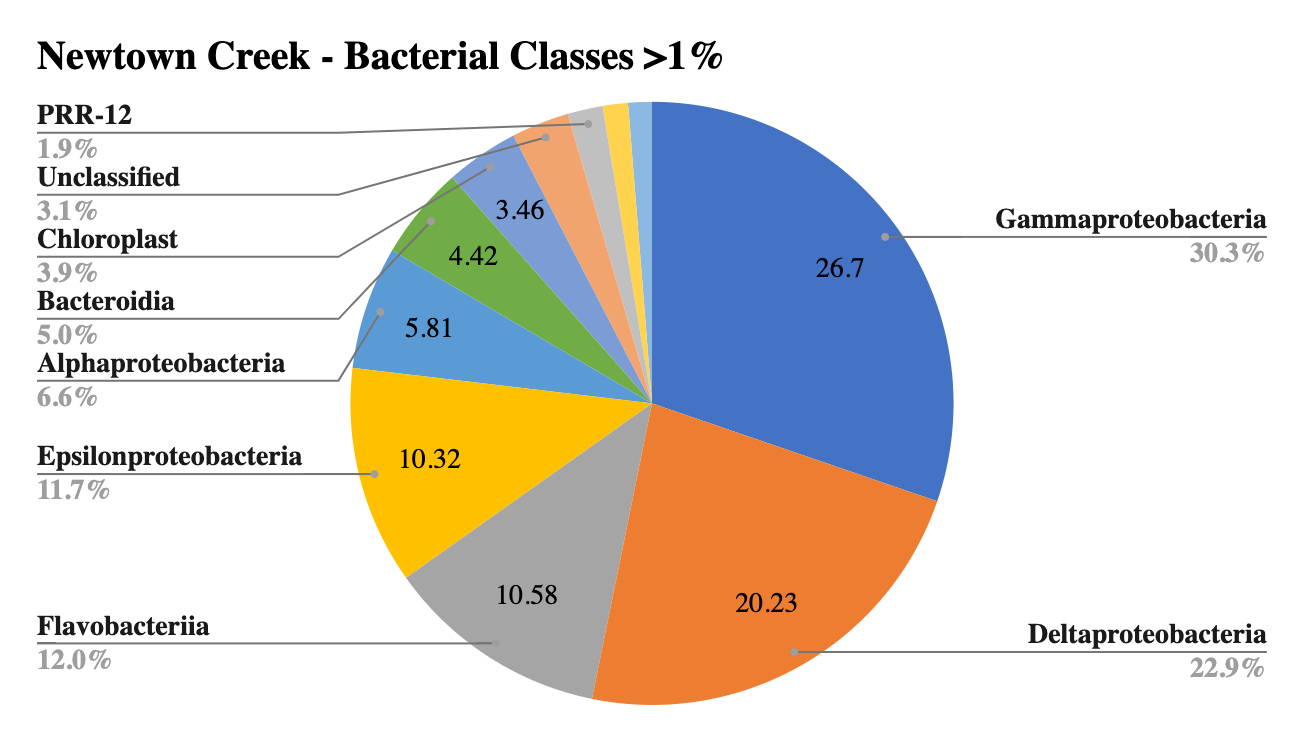
Figure 4: Bacterial diversity by class at Newtown Creek.
Environmental conditions of surrounding water indicated a range of pH, nitrate levels, salinity, temperature, phosphate, ammonium, and dissolved oxygen, reflecting the unique microenvironments of each location (Figure 5). These differing conditions were also evident by the different macroorganisms that were present such as rockweed (Ascophyllum nodosum), cordgrass (Spartina alterniflora), sea lettuce (Ulva lactuca), succulent seaweed (Sarcodiotheca gaudichaudii) and the Atlantic ribbed mussel (Geukensia demissa).
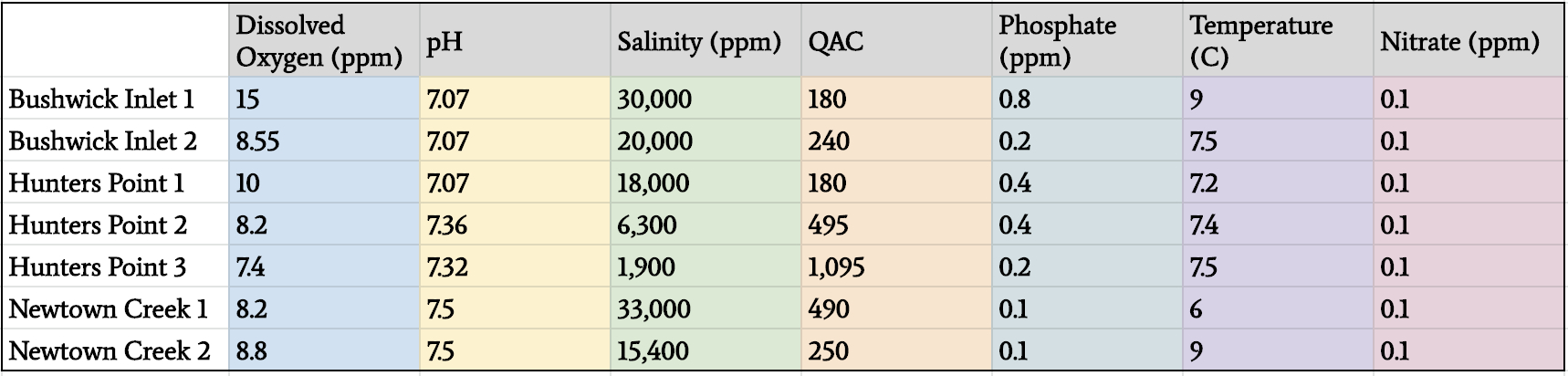
Figure 5: Results of the environmental and ancillary measurements.
By measuring the microbial diversity present in the sediment and water, we can see that a wide range of biogeochemical processes and conditions are present at each site based on the number, type and characteristics of the bacterial classes detected. Bacteria and microbes respond rapidly to changes in the environment, even on a diurnal basis, such as the inundation of the tide and due to anthropogenic factors such as pollution or sea level rise (Madsen 2011, Cleary 2016). By characterizing these sites using their unique microbiomes, we can gain insights into these types of environmental processes and changes at each site. As restoration of wetland sites in urban areas is gaining increased attention and new strategies for measuring these projects are developing, our results indicate that microbial analysis can be an effective tool towards this end.
Unique to Bushwick Inlet Park was a large portion of important cellulose decomposers and remineralizers of organic materials, and bacteria that are key in sulfur respiration. These results indicate that at Site 1 there is high vegetative input both in the form of cellulose and carbon, since the oxidation of organic carbon is necessary for sulfate oxidation (Jørgensen 2019). Unique to Hunter’s Point Park were common marine sediment bacteria that anaerobically digest, suggesting lower oxygen levels in the sediment. This was surprising due to the high vegetation cover and tidal flux of the site, which would be expected to provide adequate aeration. Unique to Newtown Creek was the highest number of types of sulfur-reducing bacteria not present in other sites, indicating sulfate reduction without oxygen. This result is expected since the Newtown creek sediments are highly inorganic and composed of high levels of hydrocarbons due to historical contamination which would create a low oxygen environment. Where data was available at the genus and species level, we found that Hunter’s Point Park had the richest diversity and known species, indicating higher overall health. Similar to other studies, the most contaminated site (Newtown Creek) had the lowest microbial diversity (Mesa 2016). It is important to note, that many bacterial sequences have not yet been reported, and the majority of our samples had nucleotide sequences that were not found in the data bank used to identify specific genus and species.
While each of the three sites proved unique, more knowledge of the identification and precise roles of microbial communities in estuaries is essential for development of effective mitigation and restoration strategies for these vulnerable ecosystems. Assessing the microbiome in these sediments does give us insights into the health and biochemical processes present in each site. Additional similar studies would be worthwhile to track the evolution or progress of these bacterial communities as the land uses change.
Barbier, E.B., Hacker, S.D., Kennedy, C., Koch, E.W., Stiera, C., Silliman, B.R. (2011). The value of estuarine and coastal ecosystem services. Ecological Monographs, 81(169–193). Available online: https://doi.org/10.1890/10-1510.1
Cleary, D. F. R., Coelho, F., Oliveria, V., Gomes, N. C. M., Polonia, A. R. M. (2016). Sediment depth and habitat as predictors of the diversity and composition of sediment bacterial communities in an inter-tidal estuarine environment. Marine Ecology, 38 (2411).
Córdova-Kreylos, A. L., et. al. (2006). Diversity, Composition, and Geographical Distribution of Microbial Communities in California Salt Marsh Sediments. Applied Environmental Microbiology, 72 (5). Available online: DOI:10.1128/AEM.72.5.3357-3366.2006
de Paula, M., Jr., Costa Silva, T.A., Araújo, A.S., Lacorte, G.A. (2021). Assessments of Bacterial Community Shifts in Sediments along the Headwaters of São Francisco River, Brazil. Conservation, 1, 91–105. Available online: DOI:10.3390/conservation1020008
Western Australia Department of Water and Environmental Regulation (DWER). (2023), Estuary Sediment Quality. Available online: https://www.water.wa.gov.au/water-topics/waterways/assessing-waterway-health/estuary-sediment-quality
Jorgensen, B. B., Findlay, A. J., Pellerin, A. (2019). The Biogeochemical Sulfur Cycle of Marine Sediments. Frontiers in Microbiology, (10). Available online: https://doi.org/10.3389/fmicb.2019.00849
Madsen, E. L. (2011) Microorganisms and their roles in fundamental biogeochemical cycles. Current Opinion in Biotechnology, 22 (456–464). Available online: DOI:10.1016/j.copbio.2011.01.008
Mesa, J., Mateos-Naranjo, E., Pajuelo, E., Ángel Caviedes, M., Rodríguez-Llorente, I.D. (2016). Heavy Metal Pollution Structures Soil Bacterial Community. Water, Air & Soil Pollution, 227 (466). Available online: DOI:10.1007/s11270-016-3176-5
New York State Department of Environmental Conservation (NYS DEC), Division of Fish, Wildlife and Marine Resources, Bureau of Habitat (2014), Screening and Assessment of Contaminated Sediment.
Palinkas, C.M., et. al. 2022. Innovations in Coastline Management With Natural and Nature-Based Features (NNBF): Lessons Learned From Three Case Studies. Frontiers in Built Environment 8(2022). Available online: https://doi.org/10.3389/fbuil.2022.814180
Kennish, M.J. (2001) Coastal salt marsh systems in the U.S.: a review of anthropogenic impacts. Journal of Coastal Research, 17(731–748).
Wigand, C.; Roman, C.T.; Davey, E.; Stolt, M.; Johnson, R.; Hanson, A.; Watson, E.B.; Moran, S.B.; Cahoon, D.R.; Lynch, J.L.; Rafferty, P., (2014). Below the disappearing marshes of an urban estuary: historic nitrogen trends and soil structure. Ecological applications: A Publication of the Ecological Society of America, 24(4): 633–649.
Wood, Jaime R., et. al. (2021). Vertical distribution of prokaryotes communities and predicted metabolic pathways in New Zealand wetlands, and potential for environmental DNA indicators of wetland condition. PLoS ONE, 16. Available online: DOI:10.1371/journal.pone.0243363