by Nancy Liu-Sullivan, Caitlin Carela, Ambar Alvarenga, and Melina Turco (College of Staten Island, Biology, 2022-2024 CRSP cohort)
The work was done as a part of the CRSP program at College of Staten Island/CUNY, under the supervision of Dr. Nancy Liu-Sullivan.
This article has been published as part of the Special Edition of Ad Astra, which features the CUNY Research Scholars Program (CRSP) across The City University of New York. The issue is accessible at http://adastraletter.com/2024/crsp-special-edition/.
ABOUT THE AUTHORS

Dr. Nancy Liu-Sullivan
Dr. Nancy Liu-Sullivan, Molecular and Cellular Pharmacology, Stony Brook University School of Medicine. An expert in cancer genomics and drug discovery. Dr. Liu-Sullivan served as a senior research scientist at Memorial Sloan Kettering Cancer Center (MSKCC) before joining the biology faculty at College of Staten Island (CSI), City University of New York. Dr. Liu-Sullivan has also served as a CUNY Research Scholars Program (CRSP) mentor for 5+ years.

Ambar Alvarenga
Ambar Alvarenga is a driven and ambitious person pursuing her undergraduate studies at the College of Staten Island. She has a profound passion for medicine and filmmaking. She has chosen to major in Biology with the goal of pursuing a career as a cardiothoracic surgeon. Apart from her devotion to the medical field, Ambar has a creative spirit, which is reflected in her minor in cinema production. With an eye for storytelling through visuals, she aspires to take on a path in the realm of filmmaking.
Ambar’s dedication to academic perseverance and her thirst for knowledge led her to apply for and become a member of the CUNY (City University of New York) Research Scholars Program (CRSP). Through this program, she seeks to deepen her critical thinking and analytical skills to broaden her understanding of the foundations of science while actively engaging in research that contributes to medical knowledge.

Caitlin Carela
Caitlin Carela is a diligent scholar currently pursuing her undergraduate studies at the esteemed College of Staten Island. She is on track to earn her bachelor's degree in Biology. Ms. Carela's academic journey is marked by a profound interest in the intersection of Medicine and Oncology, particularly in the realm of cancer research. She is an active participant in the accelerated student associate degree program and a distinguished member of the CRSP Research Scholars Program, where she has demonstrated her commitment to helping advance scientific knowledge.

Melina Turco
Beyond her academic pursuits, Ms. Carela harbors a passionate aspiration to become a veterinary professional, specializing in trauma care within the dynamic environment of an animal hospital. Her dedication to academic excellence and her unwavering commitment to the welfare of animals position her as a future leader in the veterinary field.
Melina Turco is a freshman biology major at The College of Staten Island. Melina graduated from Tottenville High School in 2023 and obtained a medical assistant license from the CTE program. Melina has aspirations of becoming a Pediatric Oncologist at which Melina joined the CRSP program in hopes of learning about new cancer treatments and research with Dr. Nancy Liu- Sullivan. Melina works as a ComHab worker, at which she tries to make kids with special needs more independent and helps them join society in a smooth manner.
Glioblastoma multiforme (GBM) is the most aggressive type of brain cancer with an abysmal survival rate of merely 14.6 months and with no known cure yet. Indeed, GBM is on a short list of extremely challenging cancers to therapeutic interventions. Among multiple factors, desmoplasia, the protective wall surrounding GBM tumor microenvironment (TME) has been recognized as a major contributing factor in tumor resistance against medical interventions. Collagens which are a group of key structural proteins in the skin and connective tissues have been recognized as playing major roles in desmoplasia scaffolding in GBM. To our knowledge, however, most published literature compares collagens found in GBM versus collagens in the normal brain. More in-depth comparison in differential roles of collagens in GBM, such as the high-grade glioma (HGG) versus low-grade glioma (LGG), would provide an additional dimension to the characterization of collagens in TME. Considering how certain LGG can deteriorate to become HGG with tougher collagen-embedded desmoplasia and, therefore, enhanced resistance to therapeutics, characterizing tumor grade-associated collagen expression alterations would help expand our knowledge of collagen biology in TME with potential to identify collagen as well as collagen-associated tumor markers and new drug targets in GBM. To this end, we explored whether or not differential collagen gene expression exist in HGG versus LGG, and identified previous unappreciated roles of collagen types and their association with the overall survival of glioma patients. Here, we present our current findings as part of an ongoing project.
Gliomas are tumors that arise from glial cells, the molecular glues that play an essential supportive and protective role for neurons. The World Health Organization (WHO) has classified gliomas by severity that ranges from low-grade gliomas (LGG) consisting of Grade I and Grade II gliomas to high-grade gliomas (HGG) comprised of Grade III and Grade IV. The most aggressive type of glioma is Grade IV glioma, also termed Glioblastoma multiforme (GBM) [1]. Highly metastatic and invasive to surrounding healthy cells and tissues, patients diagnosed with GBM have poor prognosis of, on average, 3-4 months survival expectation times. Low patient overall survival is closely associated with therapeutic resistance to treatment modalities including radiation therapy, chemotherapy, targeted therapy, and immunotherapy. A major physical obstacle that contributes to GBM resistance is the protective wall termed desmoplasia, also an integral component of the tumor microenvironment. Core to this physical obstacle is a family of structural proteins called collagen. Figure 1 illustrates a typical tumor microenvironment juxtaposed with collagens that form tumor desmoplasia.
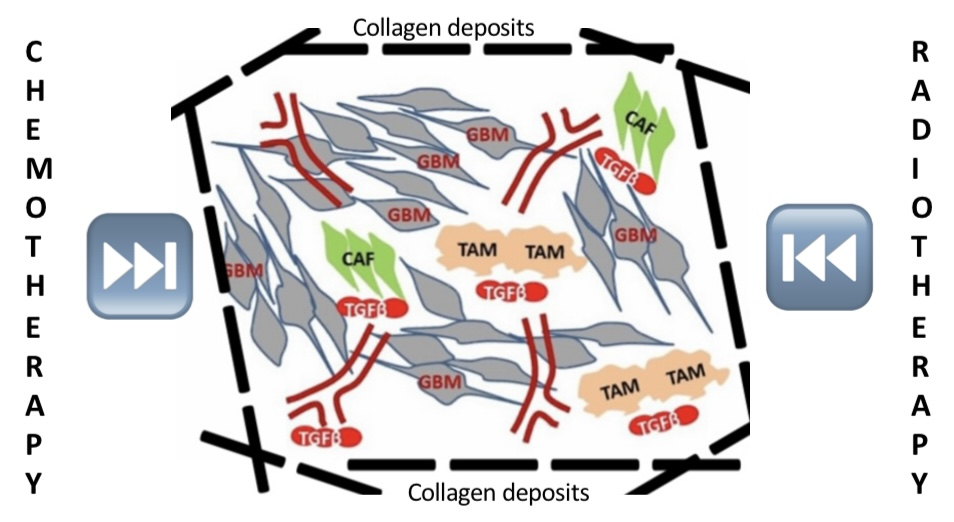
Figure 1: Shown above is the GBM tumor microenvironment (TME) illustrating the complex interplay amongst tumor proper (GBM cancer cells), collagen fibers (dark dotted lines), and other cell types including tumor-associated macrophages (TAM, beige), cancer-associated fibroblasts (CAF, green) as well as neovasculatures (red). The figure also show how collagen-rich desmoplasia interferes with radiotherapy and chemotherapy.
Indeed, levels of collagen synthesis have been reported to be elevated in GBM whereas in normal brain tissues, collagens are only produced at basal levels [2]. In particular, type III collagen A1 (Col3A1), type IV collagen A1 (Col4A1), and type V collagen A2 (Col5A1) have been observed to be elevated in GBM, a high-grade glioma. In contrast, characterization of collagen synthesis levels in low-grade gliomas such as pilocytic astrocytoma has not been given adequate attention. Pilocytic astrocytoma is a benign brain tumor that arises from astrocytes. In sharp contrast to GBM, prognosis for pilocytic astrocytoma of five-year survival is 97% [3]. Although rare, it has been reported that pilocytic astrocytoma can relapse and deteriorate to aggressive grade IV gliomas such as GBM [4]. The trajectory of collagens going from basal to highly enriched levels as pilocytic astrocytoma transforms from a slow-growing benign tumor to an aggressively proliferating and invading GBM warrants to be investigated. Here, we report ourfindings obtained from differential expression of collagen level analyses in GBM versus pilocytic astrocytoma. We also compared how collagens uniquely expressed as well as commonly expressed in GBM versus in pilocytic astrocytoma in terms of their impact on patient prognosis as reflected by overall survival.
Oncomine data query and analysis: Oncomine special edition database [5] was used for gene ranking and candidate gene selection. To ensure robustness of candidate gene selection, a stringent p value of equal or less than 0.001 and a very stringent fold change (FC) of equal to or greater than 8 were used.
Kaplan-Meier (KM) plotter overall survival analysis: Using KM Plotter private edition [6,7], we systematically analyzed all different types of collagen genes in high-grade glioma (HGG) and low-grade glioma (LGG). Gene candidates were classified by the level of expression and ranked by p value and hazard ratio (HR).
Selection of high-confidence candidate genes. For HGG selection, we focused on datasets from The Cancer Genome Atlas (TCGA) [5], an open-access program established and maintained by the National Cancer Institute (NCI) under the United States National Institute of Health (NIH). For LGG candidate gene selection, we queried datasets from a paper by Gutmann et al. [8] on the genomic characterization of pilocytic astrocytoma, a common type of low-grade benign glioma. After downloading the two sets of raw data, each set was processed by two stringent filtering criteria of retaining data points with p value equal to or less than 0.001 followed by a second filtering process of removing data points with a fold change ratio of equal to or greater than 8 of expression of experimental sample over normal brain tissues, in HGG and LGG data sets, respectively. Interestingly, COL3A1 that encodes collagen type III alpha 1 chain was found to be enriched in both HGG and LGG as a 50-fold change in LGG and 10-fold change in HGG (data not shown). We then performed a KM plotter analysis which revealed differential effects in patient overall survival for HGG versus LGG. As shown in Figure 2, COL3A1 low expression confers a survival advantage in LGG, but not in HGG.
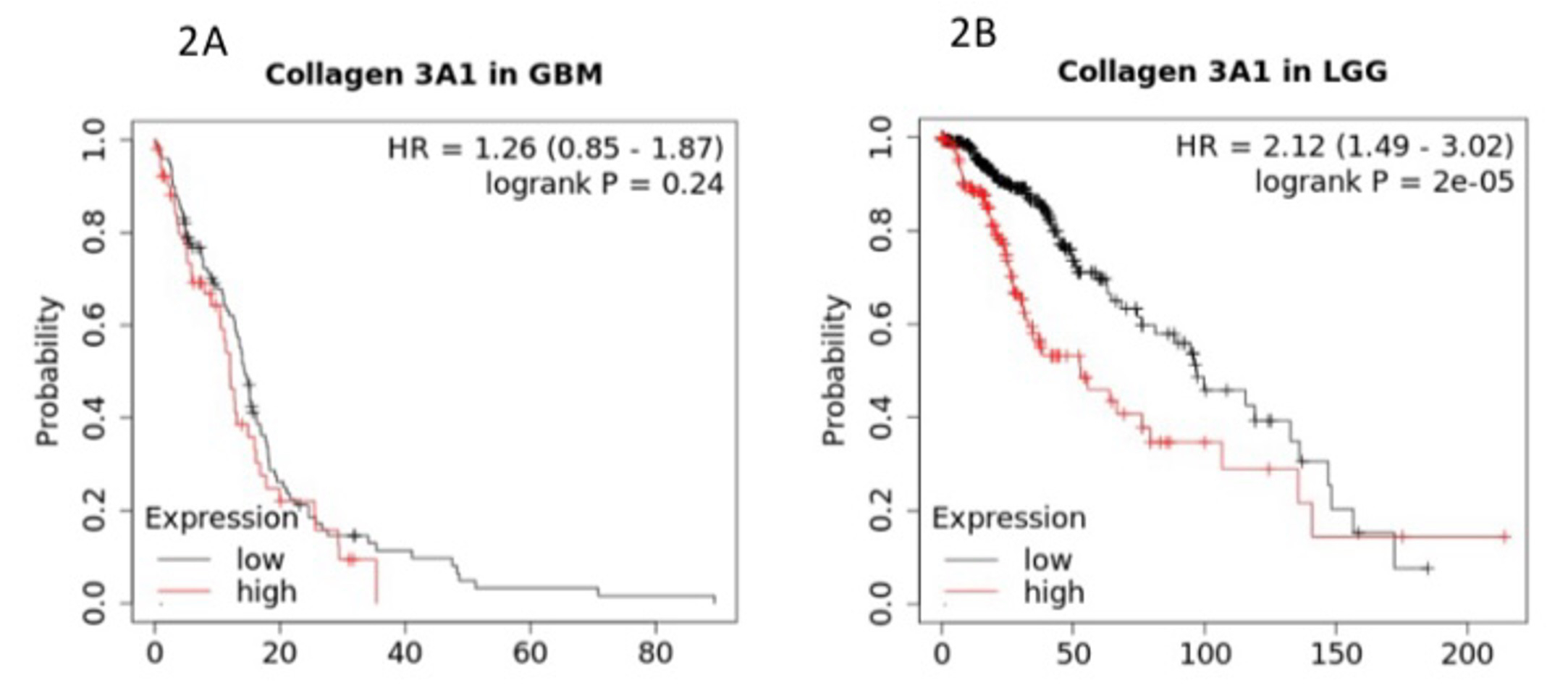
Figure 2: COL3A1 expression and overall survival in HGG (Fig. 2A) versus LGG (Fig. 2B). The Y axis represents the probability of survival as a function of time in months on the X axis.
Co-enrichment of collagen with genes essential for adhesion and structural support in HGG. In light of how neither high nor low expression of COL3A1 was found in GBM, we decided to examine the list of high-confidence genes we extracted from the GBM study. Interestingly, in addition to collagen genes, a cohort of cell adhesion-associated genes were found to be co-enriched in GBM. For example, Tenascin-C is a molecule that enhances cell adhesion and tissue remodeling in addition to promoting cell growth and metastasis in nasopharyngeal cancer [9]. An extracellular matrix protein, Periostin (POSTN) plays an important role in the formation and maintenance of bone and tooth [10]. LIMA1 with the full name of LIM domain and actin-binding protein 1 (LIMA1) is essential for maintaining integrity of cell adhesion and cell motility [11]. A Kaplan-Meier overall survival analysis in GBM in relation to the three adhesion genes is illustrated in Figure 3.
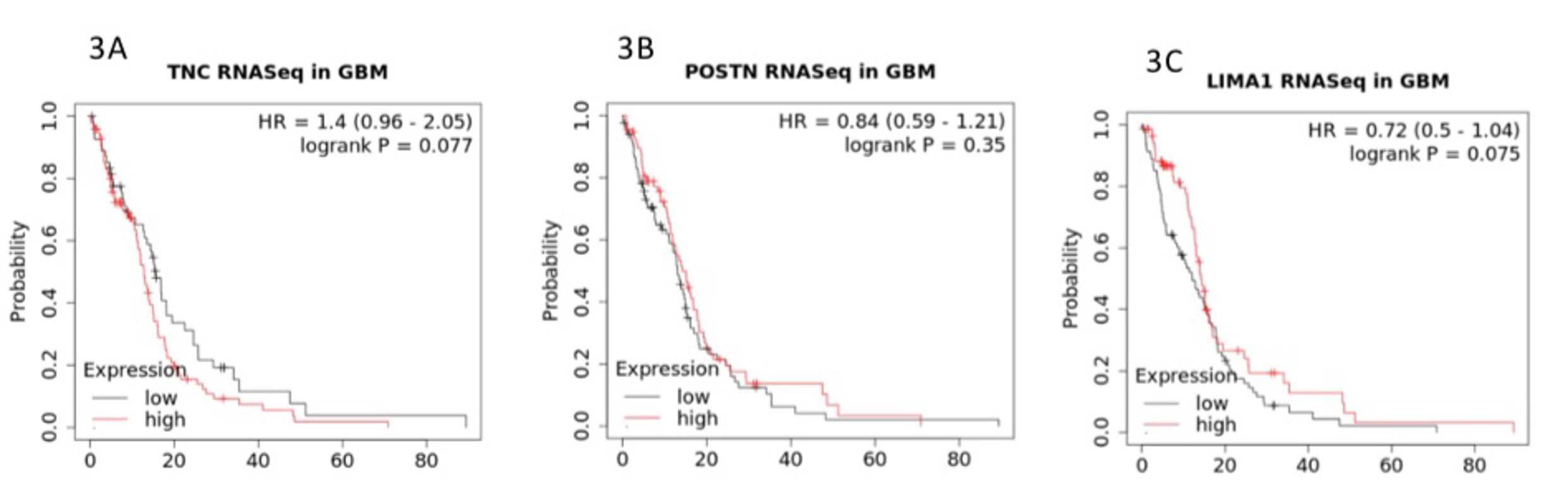
Figure 3: Co-enriched cell adhesion and cell remodeling associated genes of TNC (Fig. 3A), POSTN (Fig. 3B), and LIMA1 (Fig. 3C). The Y axis represents the probability of survival as a function of time in months on the X axis.
Co-enrichment of immune response genes. Interestingly, immune regulatory genes were also found to be co-enriched with collagens and cell adhesion genes. These genes include human leukocyte antigen DQA1 (HLA-DQA1), encoded by a class II HLA molecule, a key mediator of the adaptive immune response, reported as a pro-survival indicator in breast cancer [12]. Another highly expressed gene associated with immune response is Pentraxin 3 (PTX3), a versatile molecule that forms a three-way link amongst innate immunity, remodeling of tissues, and biogenesis of cancer [13]. The association of HLA-DQA1 and PTX3 with GBM patient overall survival is illustrated in Figure 4.
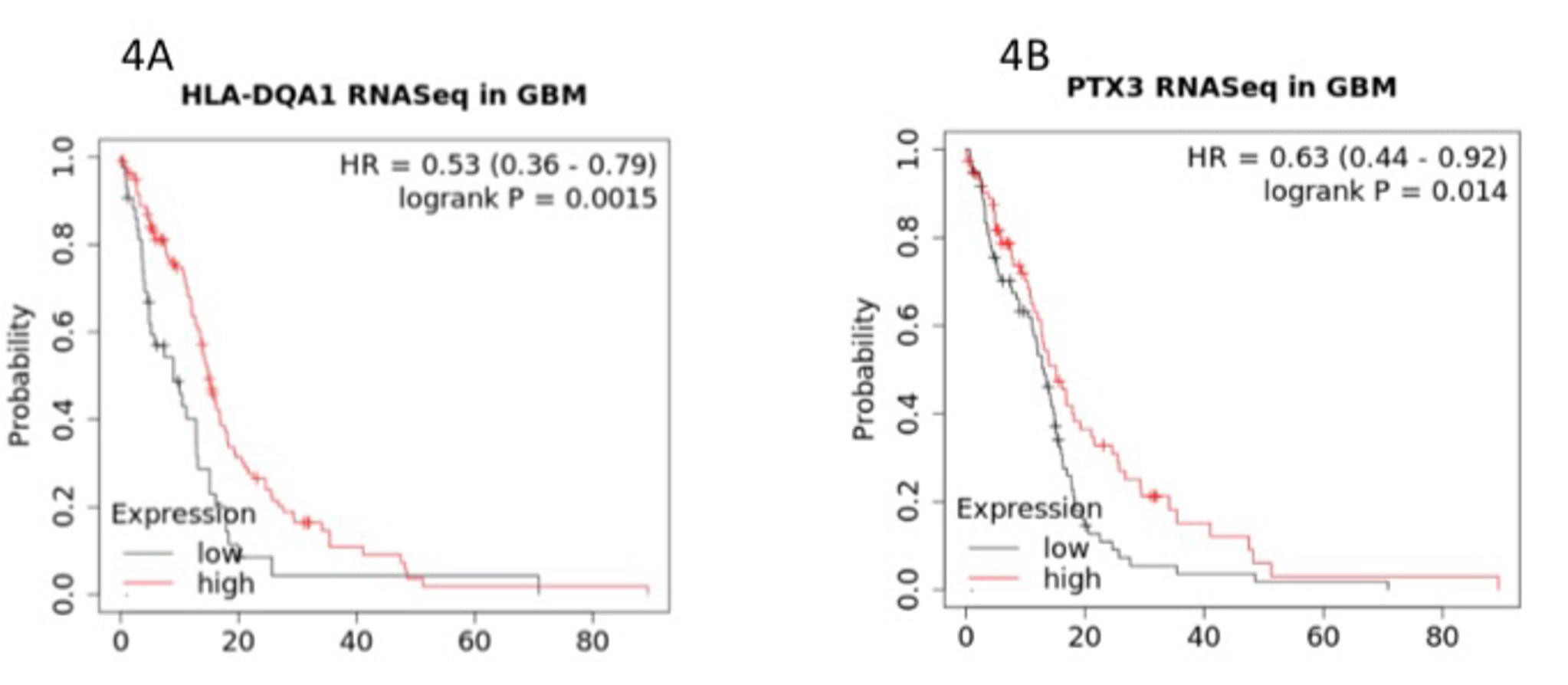
Figure 4: Co-enriched immune response-associated genes of HLA-DQA1 (Fig. 4A) and PTX3 (Fig. 4B). The Y axis represents the probability of survival as a function of time in months on the X axis.
Glioblastoma multiforme (GBM) remains an extremely challenging brain cancer as far as treatment goes, without effective medications and with a very low survival rate. Core to treatment challenges is the collagen-rich desmoplasia that serves as a protective fence around the proper tumor. Not surprisingly, the Oncomine data query and high-stringency filtering process that we carried out yielded a list of candidate genes highly enriched in GBM. To provide tumor stage-specific perspectives, we compared gene expression patterns in GBM (a type of high-grade glioma or HGG) with a low-grade glioma (LGG) termed pilocytic astrocytoma. To assess functional consequences of the highly enriched genes in HGG and LGG, we performed Kaplan-Meier overall survival analyses using a widely acknowledged analytical tool pack called Kaplan Meier Plotter (KM Plotter).
Our first finding pointed to collagen type III alpha 1 chain (COL3A1) in GBM as well as in LGG. The finding in LGG was rather striking as the COL3A1 low expression was correlated with close to 100 months of a 50% probability of overall survival in contrast to merely a little over 60 months at high level of expression. For 50% overall survival probability in HGG, however, neither high nor low levels of COL3A1 expression yielded less than 20 months. We asked the question of why COL3A1 expression deteriorated so significantly during the ascending trajectory of glioma going from low grade to high grade and whether the deterioration was associated with additional factors.
To this end, we examined the high-confidence list of extracted genes more closely using extremely stringent filtering processes and uncovered a list of enriched genes associated with processes of cell adhesion, motility, and remodeling. The three representative genes, TNC, POSTN, and LIMA1, all followed the same pattern of expression in relation to survival in that neither high nor low expression of these genes conferred survival advantages, which is consistent with the pattern of expression to survival for COL3A1. To look into the possibility of the existence of additional genes of interest negatively affecting the overall survival in GBM, we further scrutinized our high-confidence candidate gene list and observed the presence of two immune response-related genes, HLA-DQA1 and PTX3. Neither gene sets in cell adhesion or immune response were found in the LGG high-confidence candidate gene list.
Taken together, our findings point to a GBM tumor microenvironment (TME) desmoplasia scenario where collagens are at center stage, accumulating to form sturdy collagen fibers that confer a tumor-protective environment that resists therapeutic treatment. On the other hand, collagens do not exist in isolation but are instead supported by important adhesion molecules as well as by immune genes to ensure accumulation and turnover of collagens. As for immune-response gene activities, they either play a pro-inflammatory, pro-growth role or serve to help tumor cells evade immune surveillance via immune suppression. This scenario is illustrated in Figure 5. This is an ongoing project and as we gather more data, a clearer picture shall emerge.
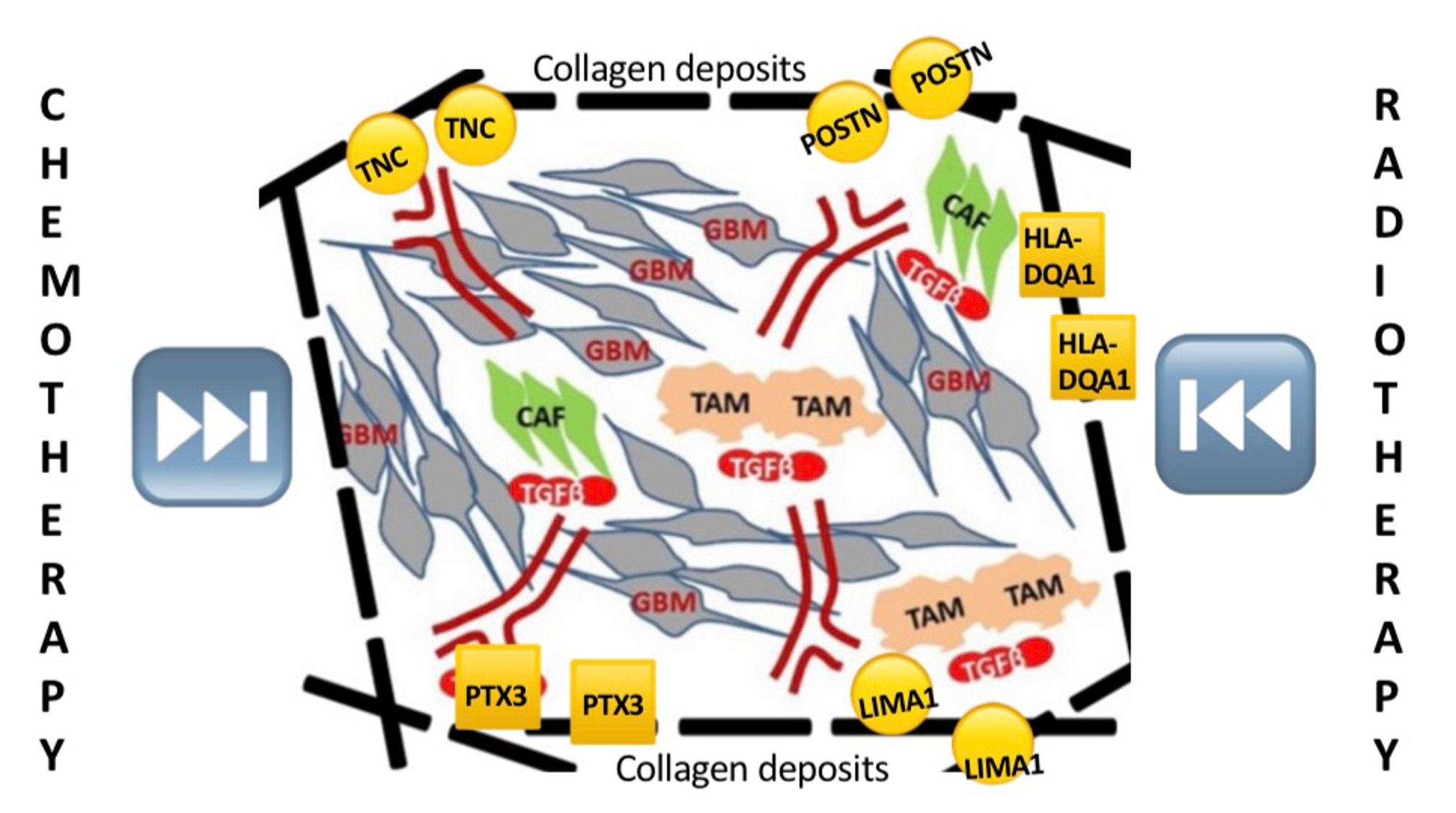
Figure 5: Schematic representation of a cooperative scenario at the tumor microenvironment that centers around collagen fibers but extending to cell adhesion, motility, and remodeling as well as immune -response molecules.
As far as we know, this is the first study focused on exploring differential roles of enriched genes in prognostics and survival of tumor grade-dependent dimensions. These and additional findings help shed additional light on the developmental progression of key genes that govern desmoplasia in GBM, which in turn can provide insights for establishing new biomarkers and potential drug targets.
The authors would like to thank Dr. Alfred Levine, PhD Director of CUNY CRSP at the College of Staten Island; Ms. Maria Ivanova, Coordinator of CUNY CRSP at the College of Staten Island; Department of Biology and Division of Science and Technology at the College of Staten Island. Our gratitude extends for Kathi Downs, Thermofisher, Ann Arbor, MI and Balázs Győrffy, MD/PhD, Department of Bioinformatics, Semmelweis University, Hungary for their scientific support.
[1] Louis DN, Perry A et al. "The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary." Acta Neuropathol, 131(6):803-20 (2016).
[2] Payne, LS. and Huang, PH. "The pathobiology of collagens in glioma." Mol. Cancer Res., 11: 1129-1140 (2014).
[3] Knight, J, De Jesus, J et al. "Pilocytic Astrocytoma - StatPearls - NCBI Bookshelf." NCBI, https://www.ncbi.nlm.nih.gov/books/NBK560614/.
[4] Rhodes DR, Yu J et al. "ONCOMINE: a cancer microarray database and integrated data-mining platform." Neoplasia, 6:1–6 (2004).
[5] "The Cancer Genome Atlas Program (TCGA)." National Cancer Institute, https://www.cancer.gov/ccg/research/genome-sequencing/tcga.
[6] Gyorffy B and Lanczky A. "Web-Based Survival Analysis Tool Tailored for Medical Research (KMplot): Development and Implementation." J. Med. Internet. Res., 23(7): e27633 (2021).
[7] http://kmplot.com/private/index.php?p=service.
[8] Gutmann, DH, Hedrick, NM et al. "Comparative Gene Expression Profile Analysis of Neurofibromatosis 1-associated and Sporadic Pilocytic Astrocytomas." Cancer Res, 62 (7): 2085–2091 (2002).
[9] Cheng, X., Li, F. et al. "Tenascin-C promotes epithelial-to-mesenchymal transition and the mTOR signaling pathway in nasopharyngeal carcinoma." https://doi.org/10.3892/ol.2021.12831.
[10] Dorafshen, S., Razmi, M. et al. "Periostin: biology and function in cancer." Cancer Cell Int., 22:315 (2022).
[11] Wang, X., Zhang, C. et al. "Characterization of LIMA1 and its emerging roles and potential therapeutic prospects in cancers." Front Immunol., https://doi.org/10.3389/fonc.2023.1115943.
[12] Zhou, JY., Xie, TT. et al. "HLA-DQA1 expression is associated with prognosis and predictable with radiomics in breast cancer." Radiat. Oncol., 18:117 (2023).
[13] Doni, A., Stravaci, M. et al. "The Long Pentraxin PTX3 as a Link Between Innate Immunity, Tissue Remodeling, and Cancer." Front Immunol., https://doi.org/10.3389/fimmu.2019.00712.