by Amena Shamia and Claudette Davis (LaGuardia CC, Biology, 2022-2023 CRSP cohort)
The work was done as a part of the CRSP program at LaGuardia Community College/CUNY, under the supervision of Dr. Claudette Davis.
This article has been published as part of the Special Edition of Ad Astra, which features the CUNY Research Scholars Program (CRSP) across The City University of New York. The issue is accessible at http://adastraletter.com/2024/crsp-special-edition/.
ABOUT THE AUTHORS

Dr. Claudette Davis
Dr. Claudette Davis is an Associate Professor in the Natural Sciences Department. She received her Ph.D. in Cell and Molecular Biology from the Graduate School & University Center, where she studied cellular senescence and apoptosis. Her post-doctoral training occurred at Howard University, where she studied the genetics of aging using Drosophila melanogaster. She began teaching at LaGuardia in Spring 2018. She currently serves as co-Director of the Biology Program and teaches Genetics, Cell Biology, and Anatomy and Physiology. In addition to STEM education research, she is interested in exploring reproduction using Drosophila melanogaster.

Amena Shamia
Amena Shamia is a biology student at LaGuardia. After earning her associate's degree in biology, she plans to pursue a bachelor's degree at a four-year university, specializing in either biotechnology or molecular biology. Currently, she is participating in research on the Drosophila melanogaster gene CG7094, supported by NIH Bridges and mentored by Dr. Claudette Davis as part of the CRSP research work. In addition to her research, she is active in campus extracurricular activities, including the Student Government Association (SGA) and the Presidents’ Society.
During my freshman year, undertaking in-depth research on bioinformatics was initially challenging due to my lack of experience in the field. However, with the foundational knowledge gained from my biology courses and the guidance of my mentor, Dr. Davis, I developed skills in navigating the genome database, interpreting genetic crosses from their coded language, and creating research posters.
Drosophila melanogaster, commonly known as the fruit fly, is a model organism used to further our knowledge about cancer genetics, aging, obesity, diabetes, and evolution. My research focuses on the evolution of genes, specifically the uncharacterized gene CG7094. CG7094 contains a protein kinase domain and belongs to the CK1 kinases (Casein Kinase 1 family). Casein kinase 1 plays a vital role in ATP production. Thus, it is likely a significant enzyme, which is also a human ortholog.
The protein sequence of D. melanogaster, CG7094, was compared with its two closest relatives, D. simulans and D. sechellia. In addition, the protein sequence of CG7094 was compared to its two older relatives, D. virilis and D. grimshawi. Our data will show that the sequences, specifically the kinase region, have undergone little evolutionary changes regardless of the environmental pressures each species encountered and the millions of years of separation.
The genome of Drosophila melanogaster is 60% homologous to that of humans. It has been estimated that nearly 75% of the genes that cause human disease are also found in the fruit fly. Thus, Drosophila can be useful in shedding light on diseases such as cancer, diabetes, and cardiovascular diseases that affect humans.
A study by Wu et al. (2020) produced 156 viable and stable transgenic flies using CRISPR-Cas9 technology. The technique disrupted phosphatase and kinase genes, which are important in signal transduction cascades. Our research focuses on one of the genes, CG7094. This gene belongs to the casein kinase (CK) gene group, and the CK domain is located on the second chromosome. The protein product is expressed in the cytoplasm and nucleus of cells. Unlike some other genes, CG7094 codes for one mRNA transcript, which will produce one polypeptide chain with 390 amino acids. Important functions of this gene product include ATP binding, signal amplification, metabolism, transcription, embryo development, apoptosis, and differentiation, all of which involve phosphorylation on acidic substrates.
The human orthologue to CG7094 is Casein Kinase 1 alpha 1(CSNK1A1). The enzyme adds phosphate groups to substrates in a process called phosphorylation. It acts as a switch for various cellular processes such as signal amplification, apoptosis, ATP binding, and more. These enzymes add phosphate groups specifically to serine and threonine amino acids. Dysfunction of CK1 can lead to DNA damage, which is one of the causes of Alzheimer's disease in humans.
We hypothesize that when speciation took place in the Drosophila genus, there were significant changes in CG7094 and its protein sequence as evolution occurred.
Flybase.org was used to obtain information about CG7094. Information on genes, protein domains, and links to the BLAST tool.
National Center for Biotechnology Information (NCBI) and Basic Local Alignment Search Tool (BLAST). Using BLAST, we found the casein kinase sequences from each species and compared them (perform BLAST) with D. melanogaster.
We started researching the selected genes in flybase.org that were uncharacterized, through which a vast amount of information could be gathered. For instance, the human orthologs, the number of RNA transcripts, the length of amino acid chains, and the functions and expression. For the gene symbol CG7094, we found that it is highly expressed in male germ line cells.
NCBI data revealed the chain of amino acids/proteins formed using CG7094 and nucleotide sequences. Using BLAST, two primitive ancestors (D. grimshawi and D. virilis) and two recent ones (D. simulans and D. sechellia) of D. melanogaster species were selected. Moreover, using the comparative sequences found in BLAST, the identities – when amino acids are the exact same, gaps – missing amino acids, and positives – similar amino acids – when each of the four species is compared with D. melanogaster was determined. In Figure 1, the BLAST result between D. melanogaster and D. simulans is shown where each letter in the chain represents an amino acid; the shaded letters correspond to different amino acids present in the species having the same functions, identities are the same amino acids and positives include both identities and non-identities. Figure 2 contains the BLAST results between D. sechellia and D. melanogaster; Figures 3 and 4 contain the BLAST results between D. melanogaster and two older species, D. virilis, and D. grimshawi.

Figure 1: BLAST of D. simulans and D. mel protein sequence. Using the BLAST result components (Identities, Positive & Gaps), we can visualize the % similarities between the two species is 97%, indicating that it is a very recent relative.

Figure 2: BLAST of D. sechillia and D. mel protein sequence. Using the BLAST result components (Identities, Positive & Gaps), we can visualize the % similarities between the two species is 94% indicating that it is a recent relative. The yellow shading represents gaps in amino acids absent in D. sechillia.
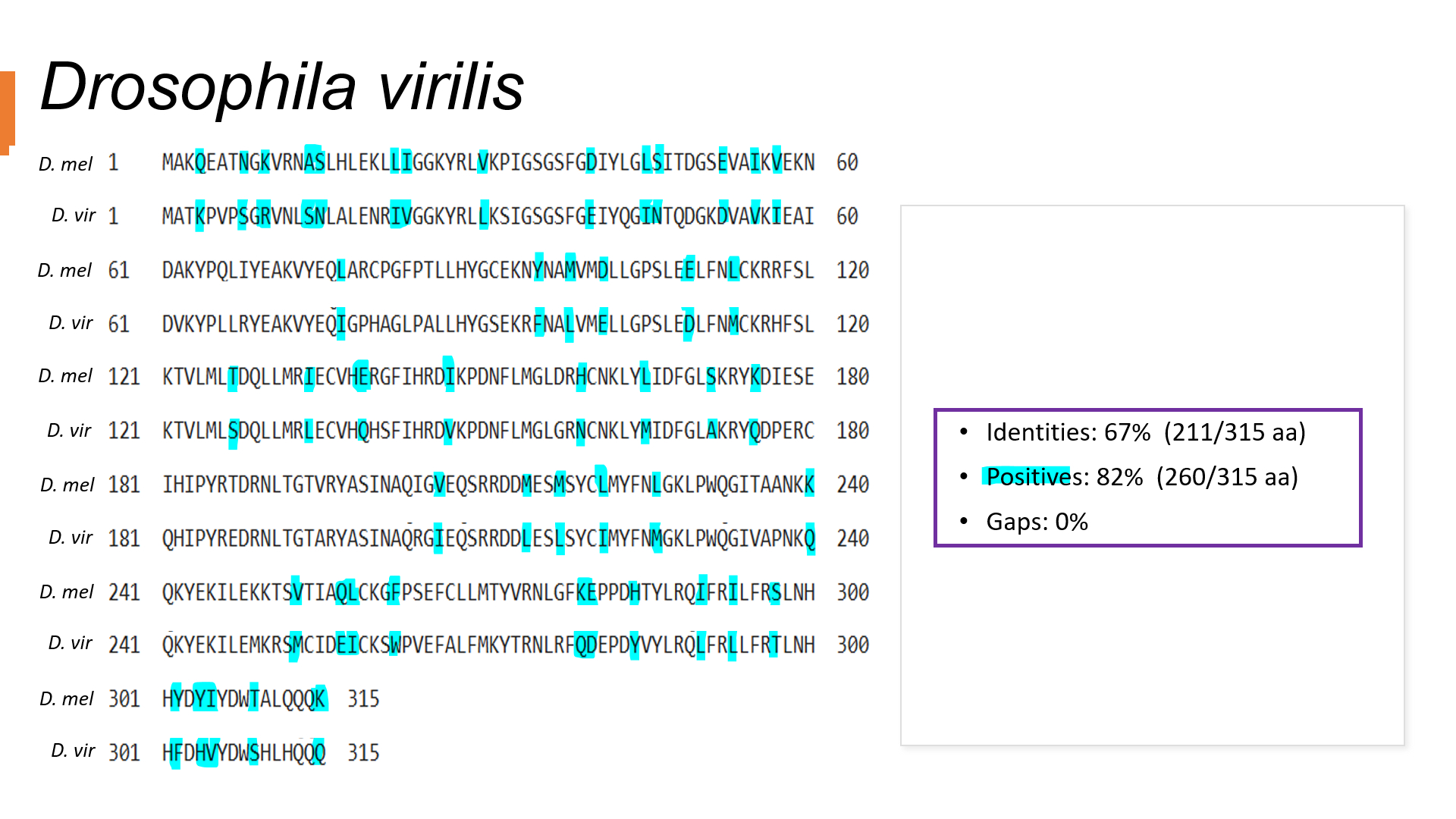
Figure 3: BLAST of D. virilis and D. mel protein sequence. Using the BLAST result components (Identities, Positive & Gaps), we can visualize the % similarities between the two species is 82%, indicating that it is a distant relative compared to others. A lower % identity indicates more differences in amino acids, so species are further apart.

Figure 4: BLAST of D. grimshawi and D. mel protein sequence. Using the BLAST result components (Identities, Positive & Gaps), we can visualize the % similarities between the two species is 81%, indicating that it is a far distant relative. Only about 65% of amino acids are found to be the same.
Using the data from the BLAST, we can conclude that D. simulans is the closest relative of D. melanogaster due to fewer changes in the protein sequence, and D. grimshawi is the oldest due to the largest change in the % positives in BLAST results.
Figure 5 below shows the cumulative relation formed during BLAST in chart form. As you move right off the graph, the percentage of positives and identities decreases, which coincides with the species being distant from melanogaster.
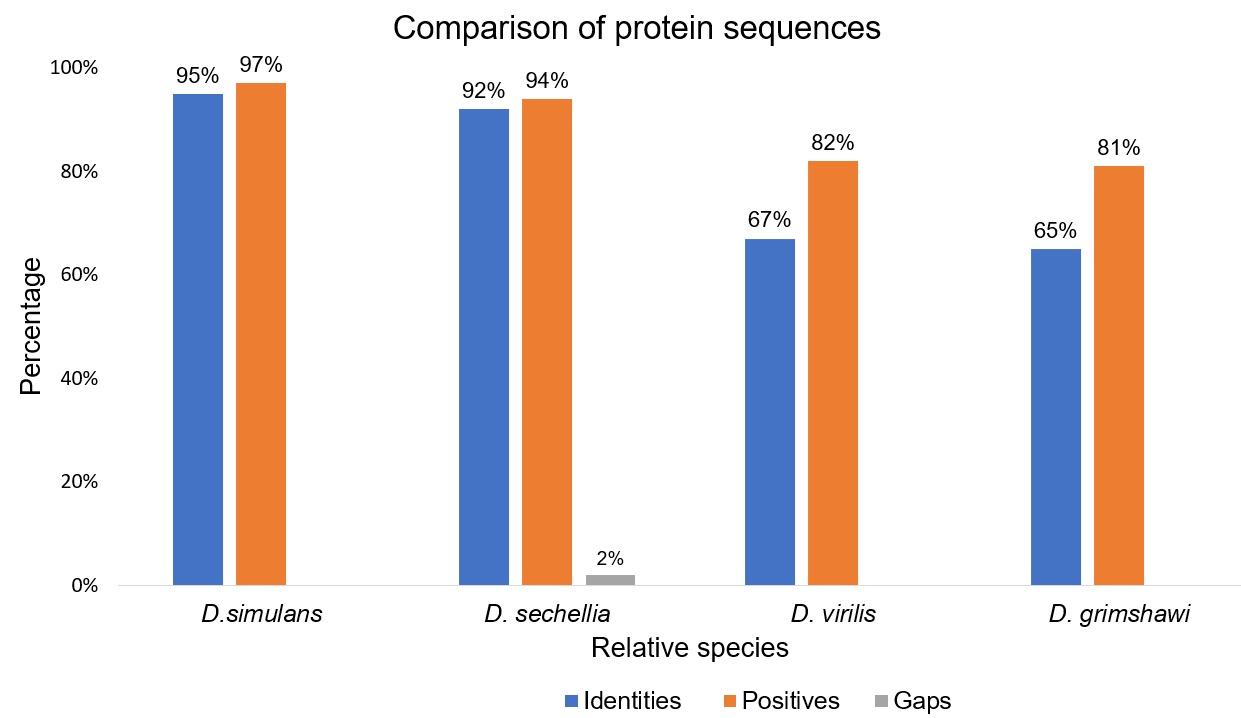
Figure 5: Comparison of protein sequences. Using the BLAST result components (Identities, Positive & Gaps), we can visualize the relationship when placed in a bar chart. This chart depicts that as the species are older, the % similarities with D.mel decreases compared to the more recent species.
The result of the research can be depicted in Figure 5. We concluded that a high percentage of amino acids were conserved over millions of years as an indication of casein kinase protein’s major usage and necessity within cell function. Based on our findings, our hypothesis was not supported, suggesting no significant changes in the protein sequence of CG7094 over time.
Additional discoveries and questions came about while conducting this research. For instance, CG7094 is highly expressed in male germ line cells. Through support from the NIH Bridges program, this project will continue to explore the role of CG7094 in reproduction by using RNAi to help determine whether CG7094 is essential for male fertility.
This research was conducted with the support of the CUNY Research Scholars Program.
[1] "FlyBase Gene Report: Dmel\CG7094." Beta.flybase.org, beta.flybase.org/reports/FBgn0032650.htm. Accessed 24 December 2023.
[2] "Interpro7-Client." Ebi.ac.uk, 2019, www.ebi.ac.uk/interpro/entry/InterPro/IPR000719/.
[3] Kapun, Martin, et al. "Drosophila Evolution over Space and Time (DEST): A New Population Genomics Resource." Molecular Biology and Evolution, vol. 38, no. 12, 2021, pp. 5782–805, https://doi.org/10.1093/molbev/msab259.
[4] Pandey, Udai Bhan, and Charles D Nichols. "Human disease models in Drosophila melanogaster and the role of the fly in therapeutic drug discovery." Pharmacological reviews vol. 63,2 (2011): 411-36. doi:10.1124/pr.110.003293.
[5] Robinson, Chloe E., et al. "Evolution of Reproductive Isolation in a Long-Term Evolution Experiment with Drosophila Melanogaster: 30 Years of Divergent Life-History Selection." Evolution, vol. 77, no. 8, 2023, pp. 1756–68, https://doi.org/10.1093/evolut/qpad098.
[6] Wu, M., Zhang, S., Wei, W., Long, L., An, S., Gao, G. (2020). "CRISPR/Cas9 mediated genetic resources for unknown kinase and phosphatase genes in Drosophila." Nature Research Scientific Reports, https://doi.org/10.1038/s41598-020-64253-4.