by ShaniaKay Williams and Bansari Patel (Queensborough CC, Biology, 2021-2022 CRSP cohort)
The work was done as a part of the CRSP program at Queensborough Community College/CUNY, under the supervision of Dr. Mangala Tawde.
This article has been published as part of the Special Edition of Ad Astra, which features the CUNY Research Scholars Program (CRSP) across The City University of New York. The issue is accessible at http://adastraletter.com/2024/crsp-special-edition/.
ABOUT THE AUTHORS

Bansari Patel
Bansari Patel is a dedicated and highly motivated student known for her exceptional work ethic. She holds an A.S. Degree in Biotechnology from Queensborough Community College and earned a place on the Dean's List for Fall 2022. Bansari also possesses a B.S. Degree in Biotechnology from Gujarat University, Ahmedabad, India, and is currently pursuing a B.A. Degree in Biological Science at Baruch College, CUNY, expected to be completed in December 2023. Her academic journey includes participation in the CUNY Research Scholar Program at QCC, where she worked on screening, isolating, and identifying new strains of Streptomyces for antimicrobial production. She gained additional research experience through the BI 554 Research Laboratory Internship at QCC, focusing on scoring liver slides under a microscope and on cell culturing. Beyond academics, Bansari has a rich volunteer background, contributing to various events in India and showcasing her commitment to community engagement and service.

ShaniaKay Williams
ShaniaKay Williams' early fascination with science led her to earn an associate degree in Natural Science, specializing in Chemistry and Biology, in Jamaica, her home country. She holds a double diploma in Agriculture Science and co-authored a food safety manual with educator Harris Dobson. At Queensborough Community College (QCC), Williams excelled in Health Science, maintaining a 4.0 GPA in her first year and applying her leadership skills as a Supplemental Instruction (SI) Leader, where she managed SI sessions and supported classroom instruction. Her role extended to judging at the American Chemical Society’s High School Research Poster Session and volunteering at the 26th Annual High School Research Poster Session. Recognized for her leadership, Williams represented her college at the Thurgood Marshall College Fund institute in Washington D.C. Consistently making the Dean's list from 2019-2023, she also made significant contributions to the CUNY Research Scholars Program (CRSP), earning two awards for her presentations. Currently pursuing a B.S. in Health Science at York College, she looks forward to graduating in June 2024.
Antibiotics are byproducts of metabolism produced by various microorganisms to compete with other microbes in the vicinity [1,2]. Many microbes evolve to produce antimicrobial compounds as a mechanism to protect themselves [1-3]. Ironically, these secondary metabolites have been lifesaving for humans to fight infectious diseases caused by pathogenic bacteria [1,3]. An important genus of bacteria Streptomyces that belongs to the family Streptomycetaceae, Class Actinomycetes, is responsible for producing a diverse range of more than half of clinically important antibiotics that are used today to treat serious infections such as Tuberculosis [3]. Industrial scale efforts are presently underway to find new strains of these soil-dwelling bacteria to generate more potent and effective antibiotics for our battle against infectious diseases [4-6]. Streptomyces include aerobic, multicellular, filamentous soil-dwelling bacteria with high Guanine + Cytosine (G+C) content in their genome. They give soil its characteristic earthy odor by producing the chemical Geosmin [7].
Antibiotic resistance is currently a serious concern in healthcare since more pathogens are becoming resistant to commonly used antibiotics due to misuse and overuse of antibiotics [1-3]. A major cause of antibiotic resistance is the acquisition of resistance genes via horizontal gene transfer among unrelated pathogens [8,9]. Although the acquisition process of resistance genes lacks complete understanding, we know that environmental microbes, including the species producing antimicrobial agents, are crucial sources of resistance transfer among species [8,9]. Antibiotic-producing bacteria harbor resistance elements for self-protection that are often clustered together with genes that code for antibiotics. (8, 9) The overuse of antibiotics is exposing common environmental bacteria to large amounts of antibiotics, which may lead to the evolution of resistance in them. Therefore, exploring antibiotic resistance occurrences in environmental microbes is critical.
Here we have isolated novel Streptomyces strains from diverse soils in New York and overseas. We are exploring the impact of these Streptomyces isolates on bacteria that commonly exist in our surroundings and study if the Streptomyces can produce novel antimicrobial products against common pathogens.
1. To isolate and characterize soil Actinomycetes from diverse geographical locations – New York and the West Indies.
2. Study the impact of soil Actinomycetes on environmental bacteria from crowded public places.
Sample Collection: We collected soil samples from diverse geographical locations in New York and in Jamaica, West Indies. In New York, we collected soil samples from Central Park, a local garden and lakeshores of Baisley Pond Park in New York City and samples from places such as Ochio Rios, Linstead and Spanish Town in Jamaica, West Indies.
Isolation of Soil Streptomyces bacteria on SFM agar: 1 gram of soil sample was mixed in 10 ml sterile distilled water and serial dilutions up to 10-5 were carried out. Then, 100 µl of the 10-5 dilution were spread on Soy Flour Mannitol (SFM) and Actinomyces Isolation Agar (AIA) plates to cultivate and isolate soil Actinomycetes. We incubated all the plates at 28-30°C for 7 to 10 days. We picked grey/white filamentous colonies with net-like mycelia for further characterization.
Making plugs of Streptomyces on SFM agar: Streptomyces isolated from SFM were re-streaked on fresh SFM plates to obtain pure culture growth of sporulated Streptomyces spp. Once fully grown, we made agar well plugs by using the back end of 1-ml plastic pipette tips. We then transferred these agar plugs to Tryptic Soy Agar (TSA) plates containing lawns of environmental isolates to study interactions amongst both groups of bacteria.
Isolation of Environmental Bacteria: We used sterile cotton swabs to collect samples from various surfaces around our environment such as- laundromat, supermarket, crowded restaurant, doctor’s office etc. The swabs were streaked on TSA plates, and single colonies were isolated and cultured on separate TSA agar plates as well as grown in liquid broth media. We used the broth cultures to test the bacteria against commonly used antibiotics. Gram staining and Microscopy were performed on all bacteria isolates to confirm their Gram stain characteristics and morphology.
Species Identification of Actinomycetes and Environmental bacteria: was performed by standard 16s rDNA barcoding technique. We used Zymo research DNA extraction kits to extract DNA from bacteria cultures and performed Polymerase Chain Reaction (PCR) to amplify the 16S ribosomal DNA gene. We then sent the amplified DNA samples for sequencing to the Genewiz, Co. (South Plainfield, NJ). We also used Biolog assay to identify select Streptomyces isolates by following their protocol (Hayward, CA).
Antibiotic Susceptibility: We tested the environmental isolates for susceptibility / resistance against many commonly used antibiotics by using Kirby Bauer assay [10].
Growth and Isolation of Soil Streptomyces: Several species of Streptomyces bacteria were successfully isolated on SFM agar and Actinomycetes Isolation agar (AIA) plates. (Fig. 1A). Individual colonies were further re-streaked on SFM agar to obtain pure cultures (Fig. 1B).
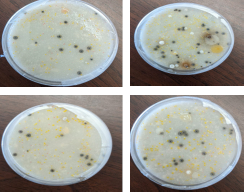
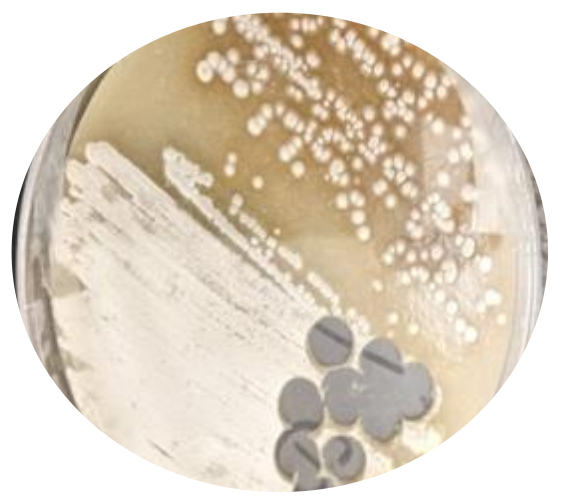
Figure 1A (Left Insret): Streptomyces colonies on SFM agar: Grey-white and other pigment colored colonies of Streptomyces were observed growing on SFM agar plates.
Figure 1B (Right Insert): Growing pure cultures of Streptomyces: Individual colonies potentially identified as Streptomyces were re-streaked on fresh SFM plates to obtain pure cultures of Streptomyces isolate/ species.
Microscopy: The Gram staining and microscopy showed that the Streptomyces isolated from soils were gram-positive filamentous bacteria (Fig. 2). Environmental samples however contained both gram-positive and gram-negative bacteria of cocci and bacilli morphology.
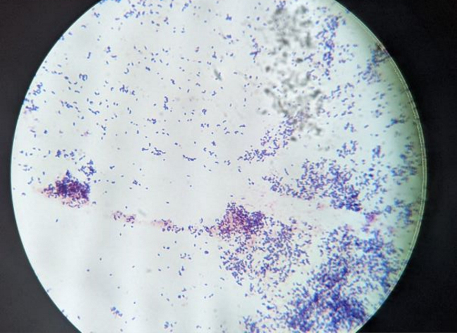
Figure 2: Gram staining of one Streptomyces isolate: Streptomyces isolates were Gram stained to observe and confirm the morphology and Gram reaction. Here is one of the isolates showing Gram-positive rod shaped bacteria at 1000X magnification.
Identification of Actinomycetes and Environmental Bacteria: The 16s rDNA sequencing revealed that the bacteria collected from a doctor's office doorknob was Bacillus thuringiensis, and one from dental office was Bacillus infantis. The Biolog assay identified one of the bacterial species from the soil samples in Jamaica WI as Microbacterium esteraromaticum; which is an actinomycete commonly found in the soil and wastewater, and as a nosocomial pathogen. Other results revealed that some of the bacteria found in the soil were microorganisms such Micrococcus lylae which belongs to Actinomycetes group that helps to keep the balance of the skin microbiota.
Testing for Antibiotic Resistance/Susceptibility: When tested for antibiotic susceptibility, results showed that the bacterium B. thuringiensis were highly resistant to commonly used antibiotics such as Cefoxitin, Penicillin, Ticarcillin, and Ceftriaxone (Fig. 3), whereas B. infantis showed resistance only to Cefoxitin antibiotic.
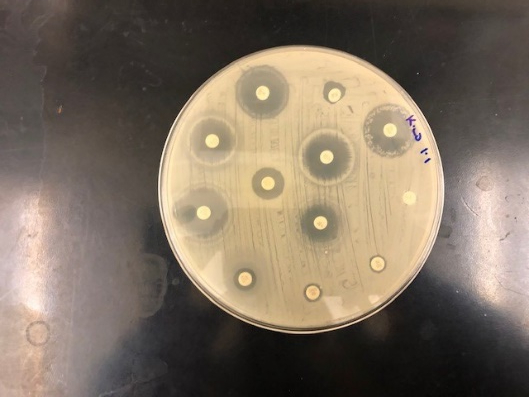
Figure 3: Kirby Baur Assay testing Antibiotic sensitivity: One of the Environmental isolate B. thuringiensis shown here indicating resistance to antibiotics such as Cefoxitin, Penicillin, Ticarcillin, and Ceftriaxone whereas sensitivity to others.
Interactions between soil Streptomyces and environmental bacteria: When co-cultured with Streptomyces, some environmental bacteria showed zones of inhibition, indicating they were either killed or prevented from growing, while others continued to grow and showed resistance to putative antimicrobial products produced by Streptomyces (Fig. 4A). This result showed that Streptomyces were producing antimicrobial substances inhibiting environmental bacteria. Further observations revealed that some Streptomyces isolates grew better in the presence of some environmental isolates. Thus, in our study, we observed both positive and negative interactions between the soil Streptomyces and the environmental bacteria. We can infer that the Streptomyces strains we collected from our soil samples did exhibit potential to produce antimicrobial metabolites.
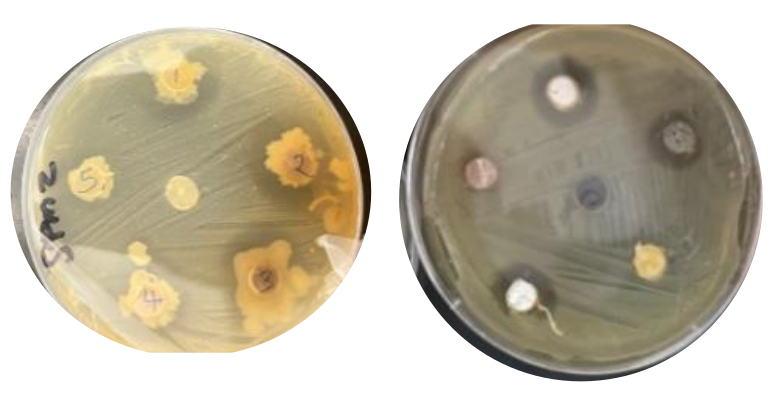
Figure 4A: Interactions between soil Streptomyces and environmental bacteria: TSA plates showing zones of inhibition against Streptomyces by environmental isolates. The two bacteria shown here were isolated from a supermarket and a laundromat.
We also studied Streptomyces for their interactions with some known bacteria that are routinely studied in Microbiology lab such as Serratia marcescens, E. coli, Proteus vulgaris etc. (Fig. 4B). Gram-negative bacteria are generally more pathogenic and less susceptible to antimicrobial products compared to Gram-positive bacteria. We observed significant size zones of inhibition produced by one Streptomyces isolate against Proteus vulgaris, which is a remarkable finding since Proteus vulgaris is a highly pathogenic Gram-negative bacterium (Fig. 4B). Thus, a soil Streptomyces exhibiting ability to secrete antimicrobial compounds that can combat this pathogen was a remarkable finding. This isolate as well as the antimicrobial metabolite(s) secreted by it need to be further studied and characterized.
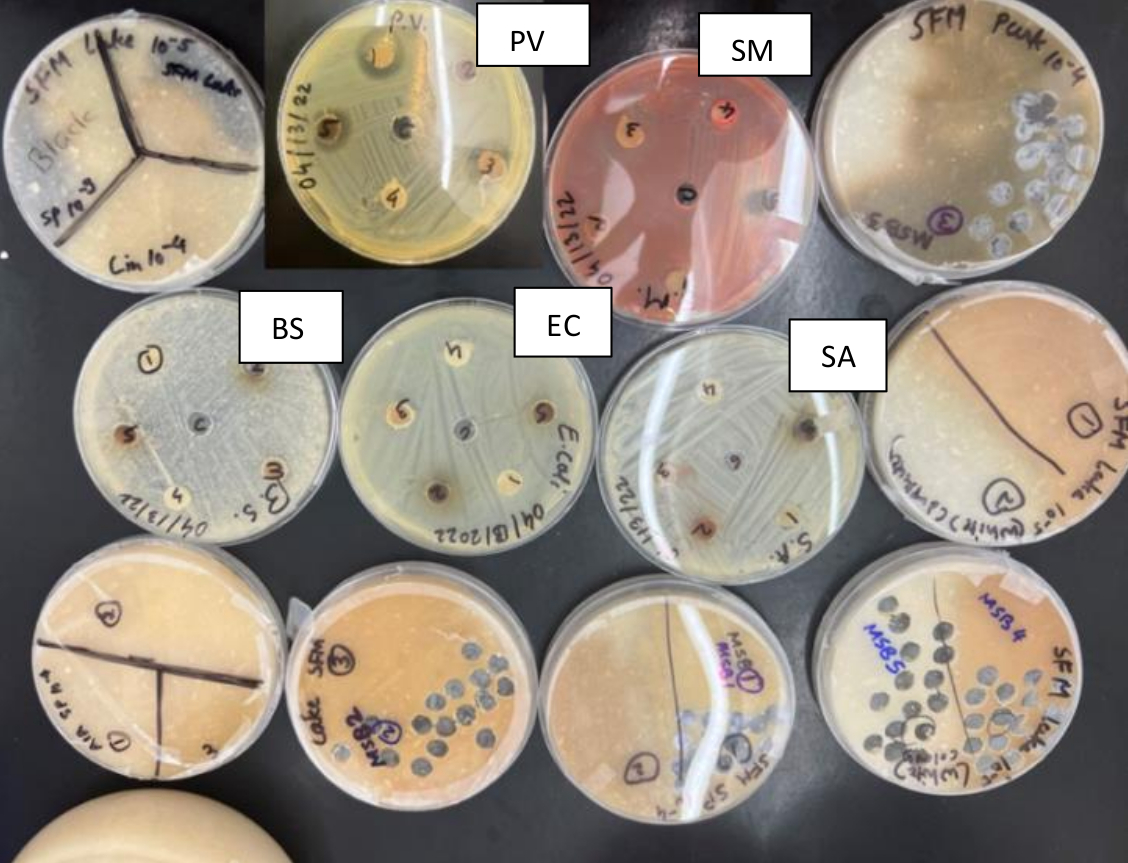
Figure 4B: Interactions between soil Streptomyces and known common bacteria: A collection of soil Streptomyces growing on SFM plates and as well as TSA plates exhibiting interaction of soil Streptomyces with some commonly known bacteria such as Proteus vulgaris (PV), Serratia marcescens (SM), Bacillus subtilis (BS), Escherichia coli (BS), and Staphylococcus aureus (SA).
Overall, we can conclude that the environmental bacteria isolated from public places were mostly found to be susceptible to antimicrobial metabolites produced by soil Streptomyces as well as antibiotics commonly used in clinical practice. Although, we were able to isolate soil Streptomyces isolates with antimicrobial potential.
[1] Danquah C. A., Minkah P. A. B., Osei P., Duah A. O., Amankwah K. B., Somuah S. O. “Antimicrobial compounds from microorganisms.” Antibiotics, 2022; 11(3).
[2] Ossai J., Khatabi B., Nybo S. E., Kharel M. K. “Renewed interests in the discovery of bioactive actinomycete metabolites driven by emerging technologies.” Journal of Applied Microbiology, 2022; 132(1):59–77.
[3] Procópio RE, Silva IR, Martins MK, Azevedo JL, Araújo JM. “Antibiotics produced by Streptomyces.” Braz J Infect Dis. 2012 Sep-Oct;16(5):466-71. doi: 10.1016/j.bjid.2012.08.014. Epub 2012 Sep 11.
[4] Atanasov A. G., Zotchev S. B., Dirsch V. M., Supuran C. T. “Natural products in drug discovery: advances and opportunities.” Nature Reviews Drug Discovery. 2021; 20(3):200–216.
[5] Srivastava N., Nandi I., Ibeyaim A., Gupta S., Sarethy I. P. “Microbial diversity of a Himalayan forest and characterization of rare actinomycetes for antimicrobial compounds.” 3 Biotech. 2019; 9(27):27–29.
[6] Selim M. S. M., Abdelhamid S. A., Mohamed S. S. “Secondary metabolites and biodiversity of actinomycetes.” Journal of Genetic Engineering and Biotechnology. 2021; 19(1).
[7] J Liato V, Aïder M. “Geosmin as a source of the earthy-musty smell in fruits, vegetables and water: Origins, impact on foods and water, and review of the removing techniques.” Chemosphere. 2017 Aug;181:9-18. doi: 10.1016/j.chemosphere.2017.04.039.
[8] Lerminiaux NA, Cameron ADS. “Horizontal transfer of antibiotic resistance genes in clinical environments.” Can J Microbiol. 2019 Jan;65(1):34-44. doi: 10.1139/cjm-2018-0275. Epub 2018 Sep 24.
[9] Jian Z, Zeng L, Xu T, Sun S, Yan S, Yang L, Huang Y, Jia J, Dou T. “Antibiotic resistance genes in bacteria: Occurrence, spread, and control.” J Basic Microbiol. 2021 Dec;61(12):1049-1070. doi: 10.1002/jobm.202100201. Epub 2021 Oct 14.
[10] Jorgensen JH, Ferraro MJ. “Antimicrobial susceptibility testing: a review of general principles and contemporary practices.” Clin Infect Dis. 2009 Dec 1;49(11):1749-55. doi: 10.1086/647952. PMID: 19857164.